Its ethanol concentration and low pH is lower than most bacteria can tolerate for growth. In most cases these microorganisms do not pose a health hazard to humans but can cause off-flavors and thus lead to the loss of entire batches and to significant.
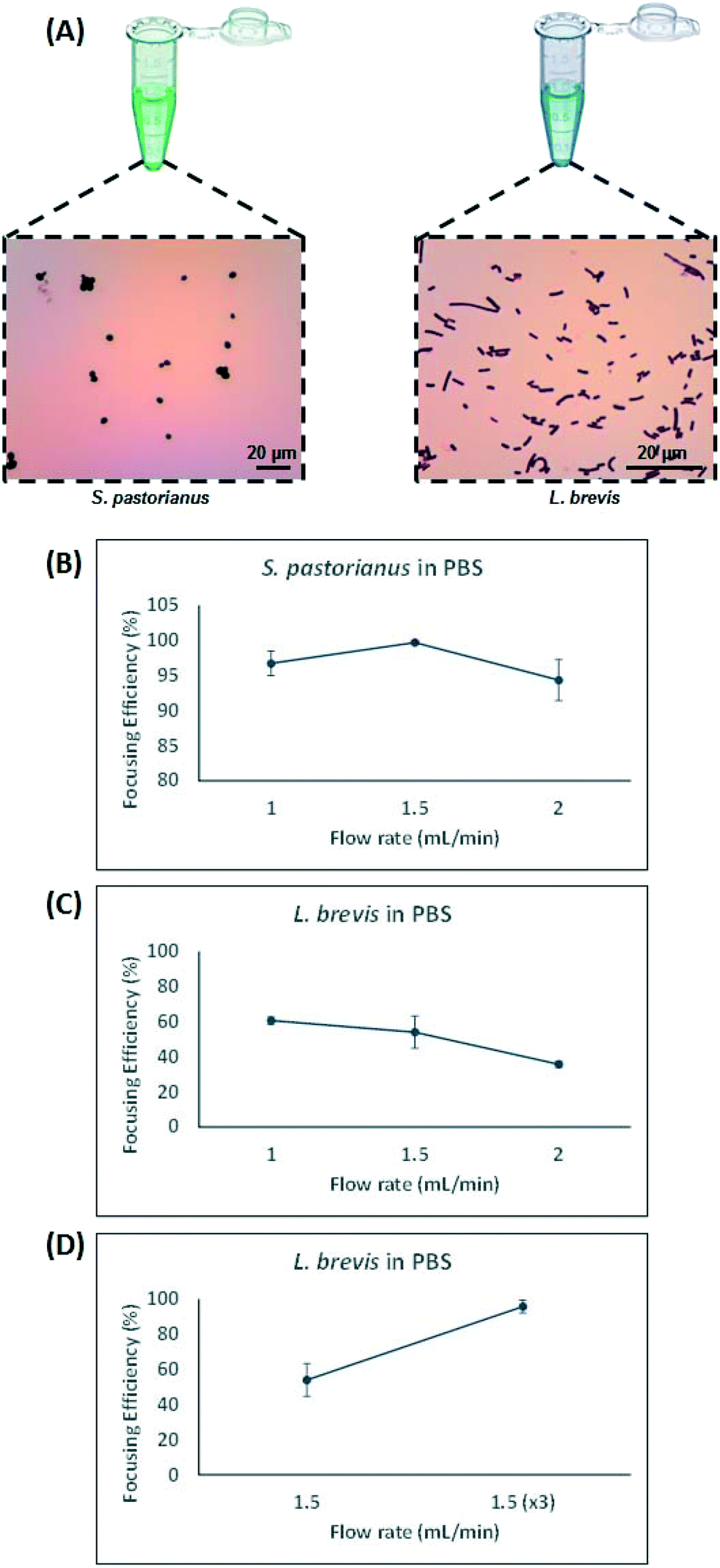
Rapid Separation And Identification Of Beer Spoilage Bacteria By Inertial Microfluidics And Maldi Tof Mass Spectrometry Lab On A Chip Rsc Publishing Doi 10 1039 C9lc00152b
Lactobacillus brevis was detected in a spoiled beer and in a commercial active dry yeast.

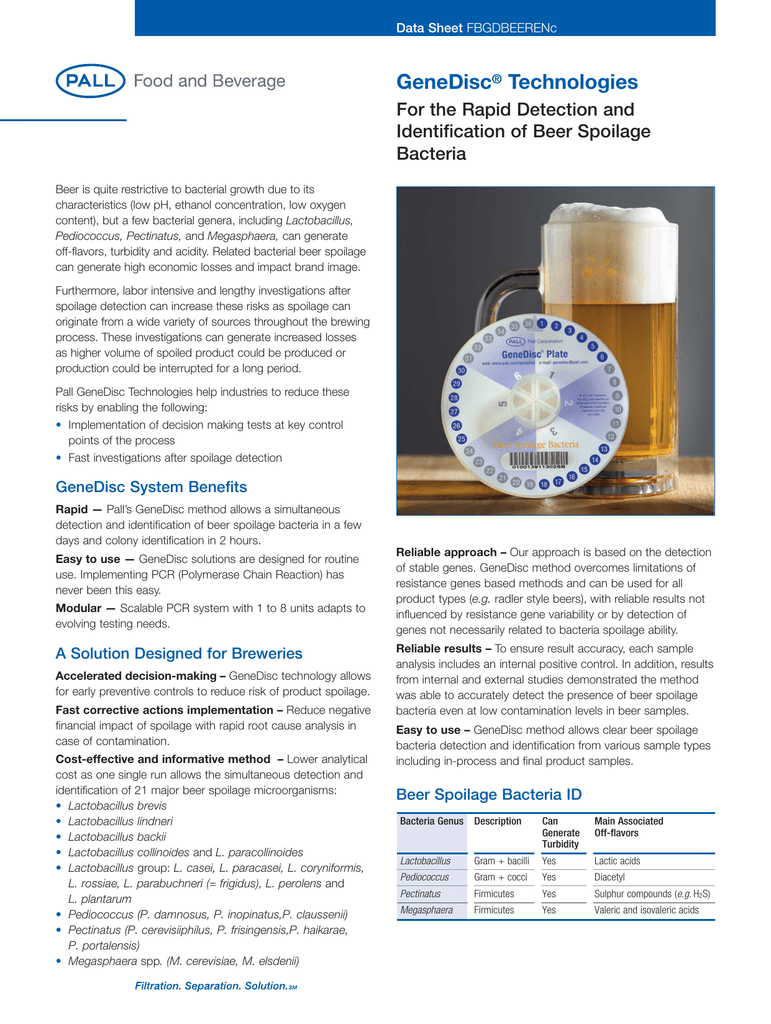
. With this method breweries can simultaneously detect and identify 21 major beer spoilage microorganisms including Lactobacillus brevis Lactobacillus lindneri Lactobacillus backii and Pediococcus. Each of these stages. Beer Spoilage Organisms Bacteria.
The presence of LAB can inhibit fermentation by the yeast and can. Beer is a poor and rather hostile environment for most microorganisms. Developing low-alcohol beers especially ones that are between 1-3 is risky due to pathogen or unwanted microbes and many brewers play safe by staying above 3.
Paradoxically it is because of beers harsh environment that bacteria are forced to use alternate internal pathways to survive and in doing so produce some of the chemicals connected with spoilage eg. Aceticacid bacteria Lpastorianus Aceticacid bacteria Zymomonasanaerobia Lpastorianus Acetomonasoxydans Pediococcus damnosus Lpastorianus Aceticacidbacteria Z. Certain endospore- forming bacteria belonging to the genus Bacillus and certain craftcoccus species craftcoccus kristinae.
And Megasphaera spp have adapted in a way that allows them to grow undisturbed even under these conditions. Were found in the brewery. Beer-spoilage microorganisms cause an increase of turbidity and unpleasant flavors in beer.
Obligate beer spoilage bacteria eg. Lactobacillus brevis was detected in a spoiled beer and in a commercial active dry yeast. 20 rows For brewing industry beer spoilage bacteria have been problematic for centuries.
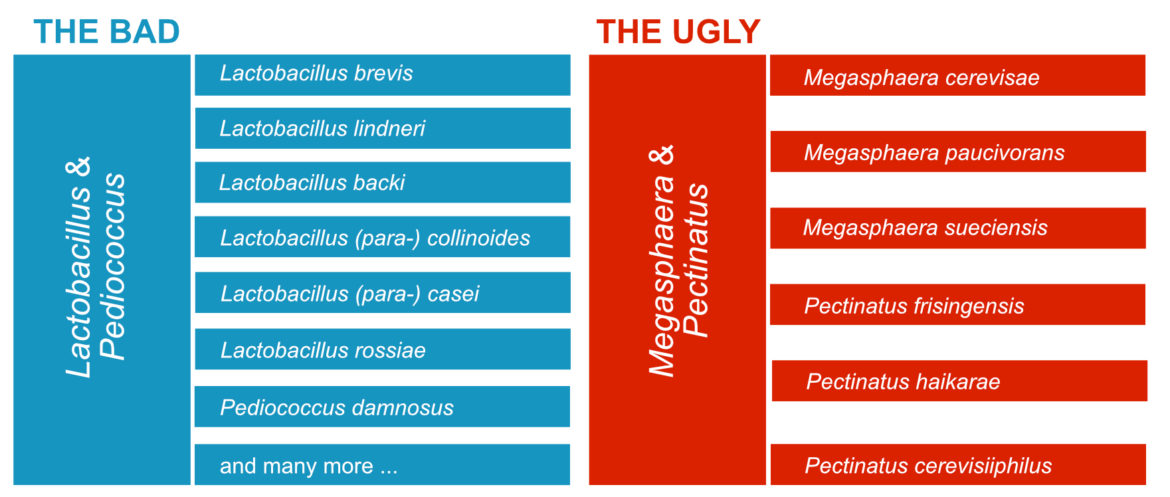
The Bacillaceae have not traditionally been considered capable of beer spoilage but four species containing the hop resistance horA gene see belowBacillus cereus Bacillus licheniformis Staphylococcus epidermidis and Paenibacillus humicushave been isolated from spoiled home-brewed beer and exhibited growth when reinoculated into beer. Other LAB species and bacteria ascribed to Staphylococcus sp Enterobaceriaceae and Acetobacter sp. They include some lactic acid bacteria such as Lactobacillus brevis Lactobacillus lindneri and Pediococcus damnosus and some Gram-negative bacteria such as Pectinatus cerevisiiphilus Pectinatus frisingensis and Megasphaera cerevisiae.
Palls GeneDisc method for beer spoilers can simultaneous detect and identify 24 of the major beer spoiling microorganisms including. Bacterial spoilage often leads to visible turbidity sediment formation acidification off-flavors and ropiness. The spoilage potential of beer spoilage bacteria is variable and is categorized according to Back 3 4 into several levels.
The important gram-negative bacteria family that have been responsible for beer spoilage are Acetic acid bacteria Zymomonas and a few members of the family Enterobacteriacea Pectinatus cerevisiiphilus and Megasphaera. A very fast method enabling monitoring of a low level of spoilers in less than 3 hours. 7 rows Three categories of beer spoilers can be distinguished.
For brewing industry beer spoilage bacteria have been problematic for centuries. Rapidly detect identify and quantify all beer spoilers right in your brewery micro lab. Anaerobia that as the brew progresses from mashing to the finished article whetherit bein cask orbottle sodoesthebacterialcontamination change in character.
Since improved process technology in modern breweries significantly reduced oxygen content in the final product the role of strictly anaerobic bacteria like Pectinatus and Megasphaera has become more prevalentDetection of these organisms which is traditionally. Beer also contains bitter hop compounds which are. Furthermore the high carbon dioxide concentration and extremely low oxygen content makes beer a near to anaerobic medium.
Nevertheless some beer spoilage bacteria like Lactobacillus spp Pediococcus spp Pectinatus spp. Chai PIKA Weihenstephan qPCR solution detects beer spoilage bacteria and wild yeasts in two hours producing clear results with Real-Time PCR. In conclusion the PCR-DGGE technique coupled with the culture-dependent method was found to be a useful tool for identifying the beer spoilage.
This highly informative method allows identification of each spoilage microorganism within 2 hours. Plating and characterizing the microbial population throughout malting revealed that some of the spoilage-associated bacteria such as Pseudomonas spp had come down compared to controls. These results are consistent with their previous findings and likely due to.
Other LAB species and bacteria ascribed to Staphylococcus sp Enterobaceriaceae and Acetobacter sp. Were found in the brewery. The quality of wine and beer suffers from the same contaminants ie lactic acid bacteria LAB and Brettanomyces.
Spoilage organisms are mainly lactic acid bacteria mostly the genera Lactobacillus and Pediococcus or they are obligate anaerobes of the species Pectinatus and Megasphaera. The main defects caused by various spoilage-causing microorganisms are. Bacteria are described as obligate beer spoilage organisms if they can grow in normal beer without an adaptation phase.
Mold species of Alternaria Cladosporium Epicoccum and Fusarium are found in spoilage beer.
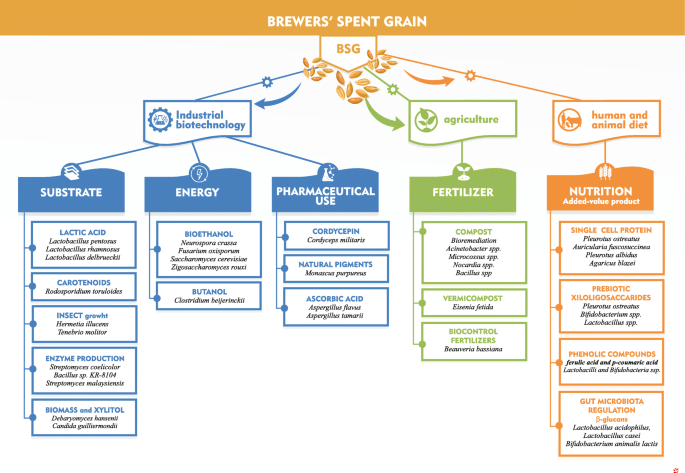
The Role Of Microorganisms On Biotransformation Of Brewers Spent Grain Springerlink
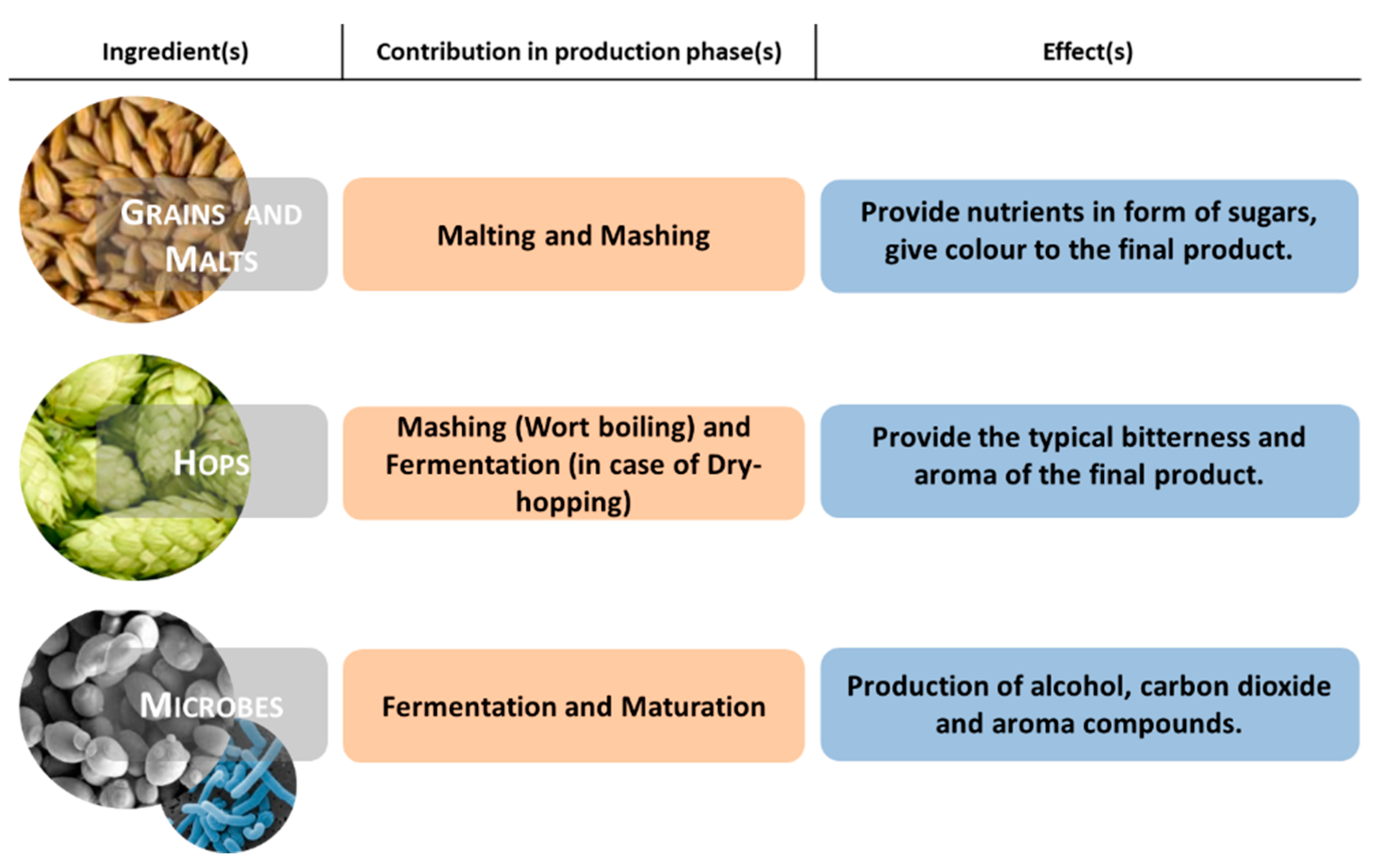
Foods Free Full Text Autochthonous Biological Resources For The Production Of Regional Craft Beers Exploring Possible Contributions Of Cereals Hops Microbes And Other Ingredients Html

Pdf Beer Spoilage Bacteria And Hop Resistance Semantic Scholar
Part 1 Beer Spoilers What Are The Major Beer Spoilers To Be Concerned About Biomerieux Industrial Microbiology

Pdf Beer Spoilage Bacteria And Hop Resistance Semantic Scholar

On The Trail Of Beer Spoilage Bacteria Food Feed Analysis

List Of Potential Beer Spoilage Microorganisms Used In This Study Download Scientific Diagram
Genedisc Technologies For The Rapid Detection And Identification Of Beer Spoilage

Pectinatus Cerevisiiphilus An Overview Sciencedirect Topics

Beer Spoilage Organisms Brewing Science

Lactic Acid Bacteria Spoilage Microbiodetection
Rapid Detection Of Beer Spoiling Bacteria Milenia Genline
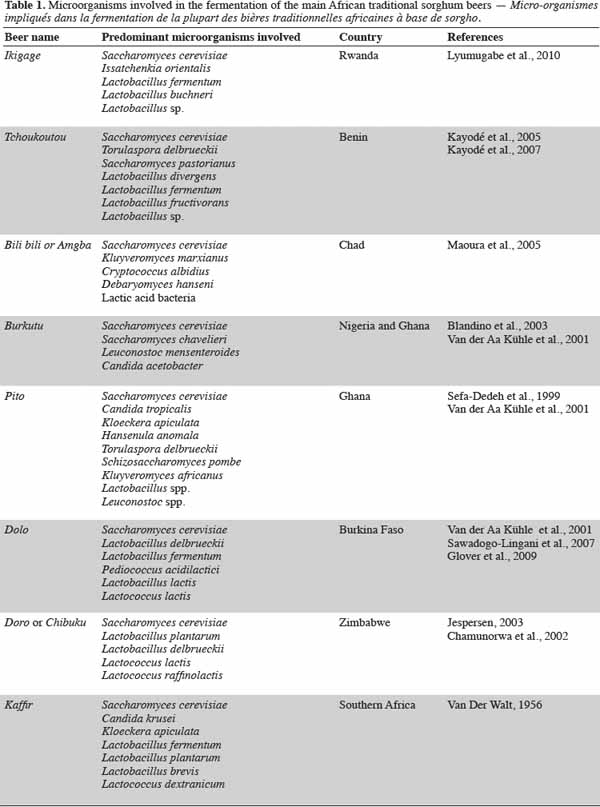
Characteristics Of African Traditional Beers Brewed With Sorghum Malt A Review Universite De Liege
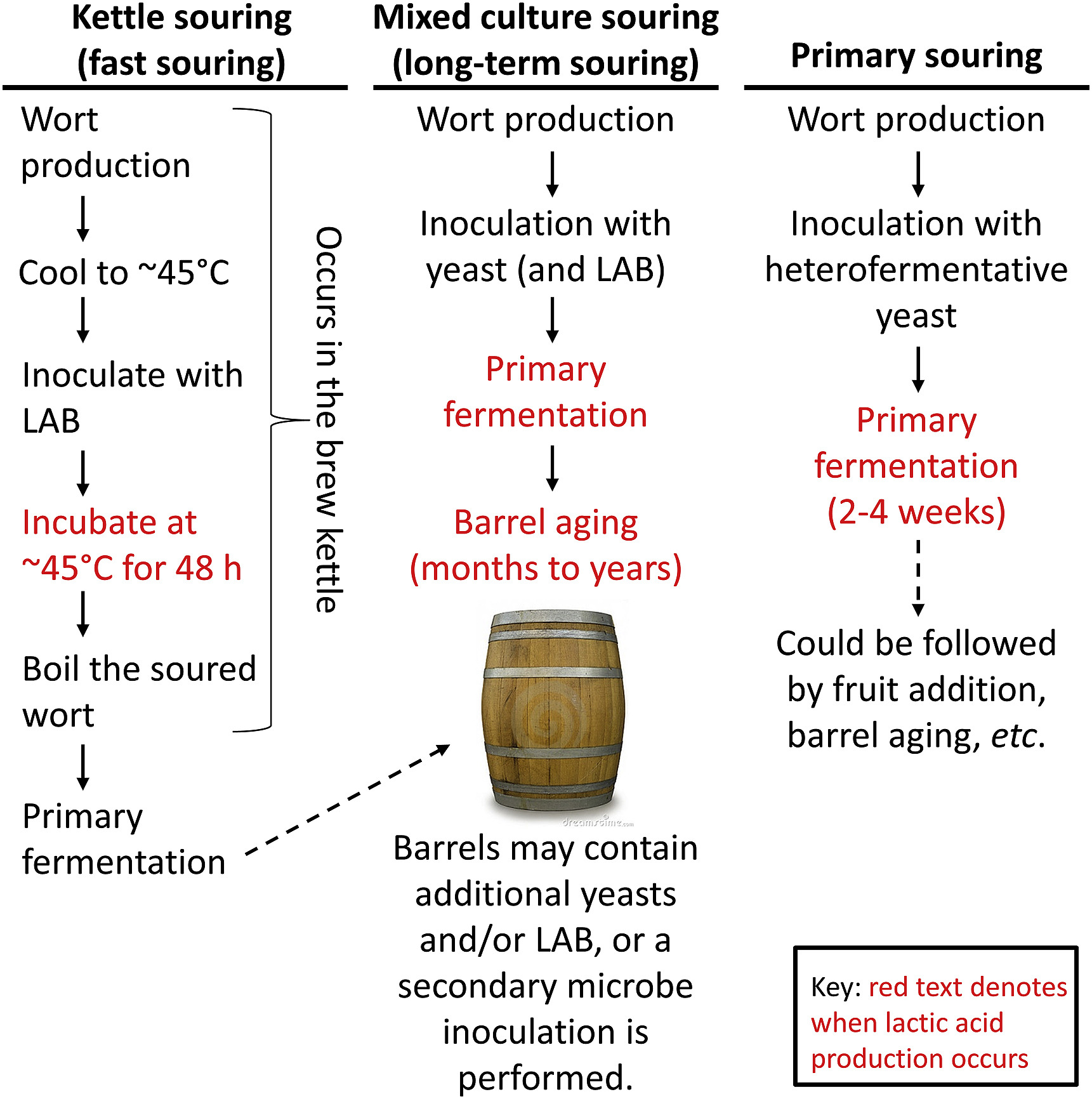
The Science Of Sour Beer Uncategorized Science Meets Food
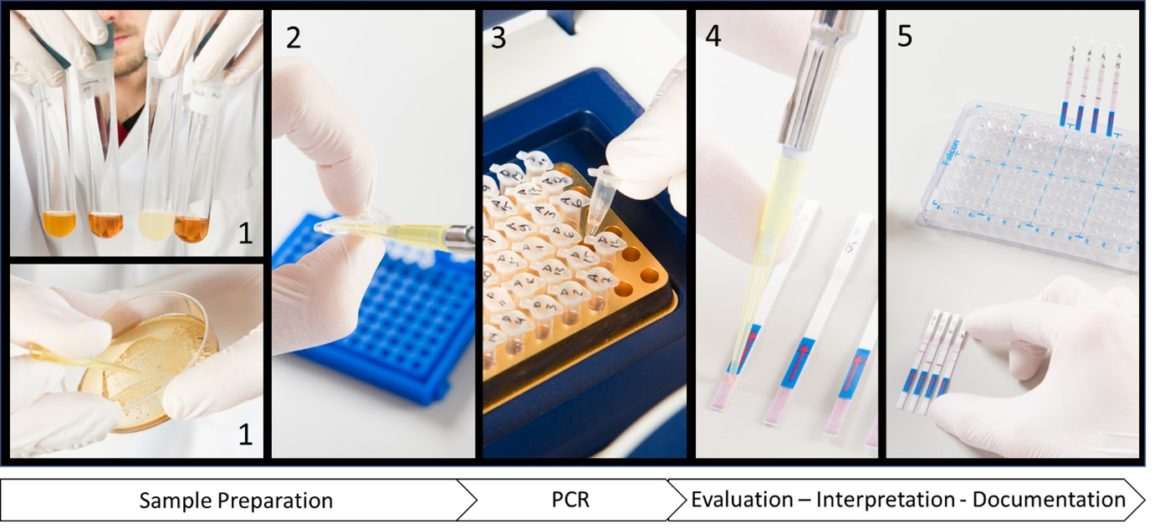
Rapid Detection Of Beer Spoiling Bacteria Milenia Genline

Culture Method For Beer Quality Control

Main Spoilage Microorganisms Of Beer Download Scientific Diagram
